
Robert P. Gunsalus, PhD
Professor Emeritus and Distinguished Research Professor
About
1602 Molecular Science Building
Microbial Cell Metabolism in Bacteria and Archaea
Affiliations:
Professor Emeritus and Distinguished Research Professor,
Department of Microbiology, Immunology & Molecular Genetics
Faculty Member:
The UCLA DOE Institute
The UCLA Molecular Biology Institute
UCLA Graduate Programs in Biosciences (GPB) Home Areas:
Immunity, Microbes & Molecular Pathogenesis GPB
Genetics & Genomics GPB
Biochemistry, Biophysics & Structural Biology GPB
UCLA Faculty Profile: https://profiles.ucla.edu/robert.gunsalus
Research Interests :
Our current research focuses on the molecular biology and physiology of anaerobic microorganisms including the syntrophic bacteria, their associated methanogenic partners plus other archaeal species. These microbes colonize a wide range of habitats on Earth and possess the ability to adapt and grow over a wide range of environmental conditions (e.g., substrate type, temperature, pH, salinity). We are examining how their metabolism, energy harvesting and associated cell envelope structures adapt to these changes using a variety of genetic, molecular and biochemical approaches.
1) Genomics of the Syntrophic Bacteria and the Methanogenic Archaea. The syntrophic group of bacteria along with their archaeal methanogen partners are expert in the recycling of biomass in most anaerobic environments. Little yet is known about their metabolism and ability to adapt to changing conditions. Using genomic, transcriptomic and proteomic approaches we are elucidating their core metabolic pathways for carbon utilization and energy harvesting. The longer-term goal is to generate predictive metabolic models that describe the co-operative partnerships between these syntrophic and methanogenic groups of microbes.
Metabolic reconstruction of Syntrophomonas wolfei


2) Microbial Cell Envelopes. We also study the composition, structure and function of archaeal cell envelopes. The majority of these have a protein coat rather than a peptidoglycan layer that surrounds the cell membrane to protect it from the environment outside. The coat or S-layer proteins are among the most abundant in the cell although their properties and structures remain poorly understood.
We have identified the major S-layer envelope proteins in several archaeal methanogenic strains including those in the genera, Methanospirillum,Methanosaeta and Methanosarcina. Following S-layer protein synthesis, post-translational modification and membrane export, they are assembled into a continuous outermost layer. Current work focuses on the analysis of their structures along with the associated cell appendages (flagella and pili). It provides new insight regarding the ability of these archaea cell types to adapt to diverse growth conditions.
Cryo EM tomogram of a Methanospirillium hungatei cell flagellum structure
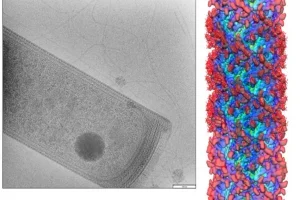
(images by Hui Wang and Hong Zhou)
Cryo EM of M. hungatei and its outer sheath layer:
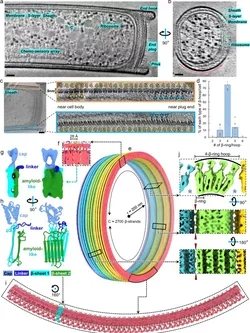
(image by Nicole Poweleit and Hong Zhou)
Biography:
Dr. Gunsalus is currently a Distinguished Research Professor in the Department of Microbiology, Immunology, and Molecular Genetics at the University of California, Los Angeles within the Schools of Life Sciences and Medicine. He obtained B.S. degrees is in Microbiology and in Chemistry from the South Dakota State University and went on to do his PhD studies in Microbial Biochemistry at the University of Illinois at Urbana Champaign working with Dr. Ralph S. Wolfe. Prior to joining the faculty at UCLA, he did postdoctoral work at Stanford University in Palo Alto in the areas of Molecular Biology with Dr. Charles Yanofsky and Dr. Robert D. Simoni.
Publications:
Hui Wang, Jiayan Zhang, Daniel Toso, Shiqing Liao, Farzaneh Sedighian, Robert P. Gunslaus and Z. Hong Zhou. 2023. Hierarchical organization and assembly of the archaeal cell sheath from an amyloid-like protein. Nat. Commun. 14:6720 PMID 37872154
Muroski, J.M,, Fu, J.Y, Nguyen, H.,H., Wofford, N.Q, Mouttaki, H, James, K.L, McInerney, M.J., Gunsalus, R.P., Loo, J.A., Ogorzalek Loo, R.R. 2022.The acyl-proteome of Syntrophus aciditrophicus reveals metabolic relationships in benzoate degradation. Molec. & Cellular Proteomics. 21: 4, 100215.
Fu, JY, Muroski, JM, Arbing, MA, Salguero, JA, Wofford, NQ, McInerney, MJ, Gunsalus, RP, Ogorzalek Loo, RR, and J.A. Loo. 2022. Dynamic acylome reveals metabolite driven modifications in Syntrophomonas wolfei. Front Microbiol 2022 Nov 7;13:1018220.
James, K.L., Kung, J.W., Crable, R.R., Mouttaki, M., Sieber, J.R., Nguyen, H.H., Yang, Y., Xie, Y., Erde, J., Wofford, N.Q., Karr, E.A., Loo, J.A., Ogorzalek Loo, R.R., Gunsalus, R.P. and M.J. McInerney. 2019. Syntrophus aciditrophicus uses the same enzymes in a reversible manner to degrade and synthesize aromatic and alicyclic acids. Environmental Microbiology. 21:1833-1846.
Lauren E. Cook, L.E., S. Gang, A. Ihlan, M. Maune, R.S. Tanner, M.J. McInerney, G. Weinstock, E.A. Lobos and R.P. Gunsalus. 2018. Genome sequence of Acetomicrobium hydrogeniformans OS1. Genome Announc. 2018 Jun; 6(26): e00581-18. Published online 2018 Jun 28. doi: 10.1128/genomeA.00581-18
Briege, A., C.M. Oikonomou, Y-W Chang, Kjaer,A., A.N. Huang, K.W. Kim, D. Ghosal, D. Kenney, R.O.O. Loo, R.P. Gunsalus, and Grant J. Jensen. 2017. “Morphology of the archaellar motor and associated cytoplasmic cone in Thermococcus kodakaraensis. EMBO Rep. Sep;18(9):1660-1670.
Poweleit, N., P. Ge, H.H. Nguyen, R.R. Ogorzalek Loo, R. Gunsalus, and Z.H. Zhou. CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pilus. Nature Microbiology. 2016. 2: 16264.
Gunsalus, R.P., L.E. Cook, B. Crable, L. Rohlin, E. McDonald, H. Mouttaki, J.R. Sieber, N. Poweleit, Z.H. Zhou, A.L. Lapidus H.E. Daligault, M. Land, P. Gilna, N. Ivanova, N. Kyrpides, D.E. Culley and M.J. McInerney. Complete genome sequence of Methanospirillum hungatei type strain JF1. Standards in Genomic Sciences. 2016; 11: 2.
Toso D.B, M.J. Javed, E. Czornyj, R.P. Gunsalus, and Z.H. Zhou. Discovery and Characterization of Iron Sulfide and Polyphosphate Bodies Coexisting in Archaeoglobus fulgidus Cells. Archaea. 2016. 2016: 4706532.
Arbing, M.A., S. Chan, A. Shin, T. Phan, C.J. Ahn, L. Rohlin, and R.P. Gunsalus. Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans. Proceedings of the National Academy of Sciences of the United States of America. 2012; 109: 11812-7.
Sieber, J.R. M.J. McInerney and R.P. Gunsalus. Genomic Insights into Syntrophy: The Paradigm for Anaerobic Metabolic Cooperation. Annual Review of Microbiology. 2012. 109(29): 429-452.
McInerney, M.J. L. Rohlin, H. Mouttaki, U.M. Kim. R.S. Krupp, L. Rios-Hernandez, J.R. Sieber, C.G. Struchtemeyer, A. Bhattacharyya, J.W. Campbell, and R.P. Gunsalus. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proceedings of the National Academy of Sciences of the United States of America. 2007; 104: 7600-5.
Lab Members:
Ethan Humm – Research Assistant
Lauren Cook – Post doc
Miisa Makela – Undergraduate Researcher
Alex Fiorito – Undergraduate Researcher
Mordecai Wesenachin – Undergraduate Researcher
Links:
UCLA Faculty Profile: https://profiles.ucla.edu/robert.gunsalus
UCLA Molecular Biology Institute: http://www.mbi.ucla.edu/
UCLA DOE Institute of Genomics and Proteomics: http://www.doe-mbi.ucla.edu/
EcoCyc – the Escherichia coli protein gene database: http://ecocyc.org/
Contact:
Dr. Robert Gunsalus
Department of Microbiology, Immunology, and Molecular Genetics
UCLA
609 Charles E Young Dr S
1602 Molecular Science Building
Los Angeles, CA 90095
(310) 206-8201 office
robg@microbio.ucla.edu